Bordetella pertussis mechanosensing system contributing to bacterial colonization (Horiguchi Lab, in Sci. Adv.)
Horiguchi et al. demonstrate that Bordetella pertussis senses the host cells through flagella and upregulates a small RNA (sRNA) contributing to bacterial infection.
The Gram-negative bacterium Bordetella pertussis causes a highly contagious respiratory disease called pertussis (whooping cough) that is characterized by severe and uncontrollable coughing. B. pertussis produces multiple virulence factors, including adhesins and toxins, at appropriate periods in the course of infection. The expression of these virulence factors was previously considered to be largely regulated by the BvgAS two-component system, which is activated when the bacteria invade the host; however, recent studies indicated that the profiles of BvgAS-regulated genes differ significantly between in vitro and in vivo conditions, suggesting a more complex network between BvgAS and other systems to regulate gene expression. Recently, we focused on the small RNA (sRNA) of the bacteria as another virulence regulator and identified nine distinct sRNAs designated Bpr (B. pertussis sRNA) 1-9 that were highly transcribed upon tracheal colonization in mice. Moreover, the expression of eight sRNAs was not regulated by the BvgAS system (Hiramatsu et al., Microbiol Immunol. 64:469-75, 2020). These results imply that these sRNAs are up-regulated in response to environmental cues within the host in a BvgAS-independent manner and participate in downstream genes that support bacterial infection.
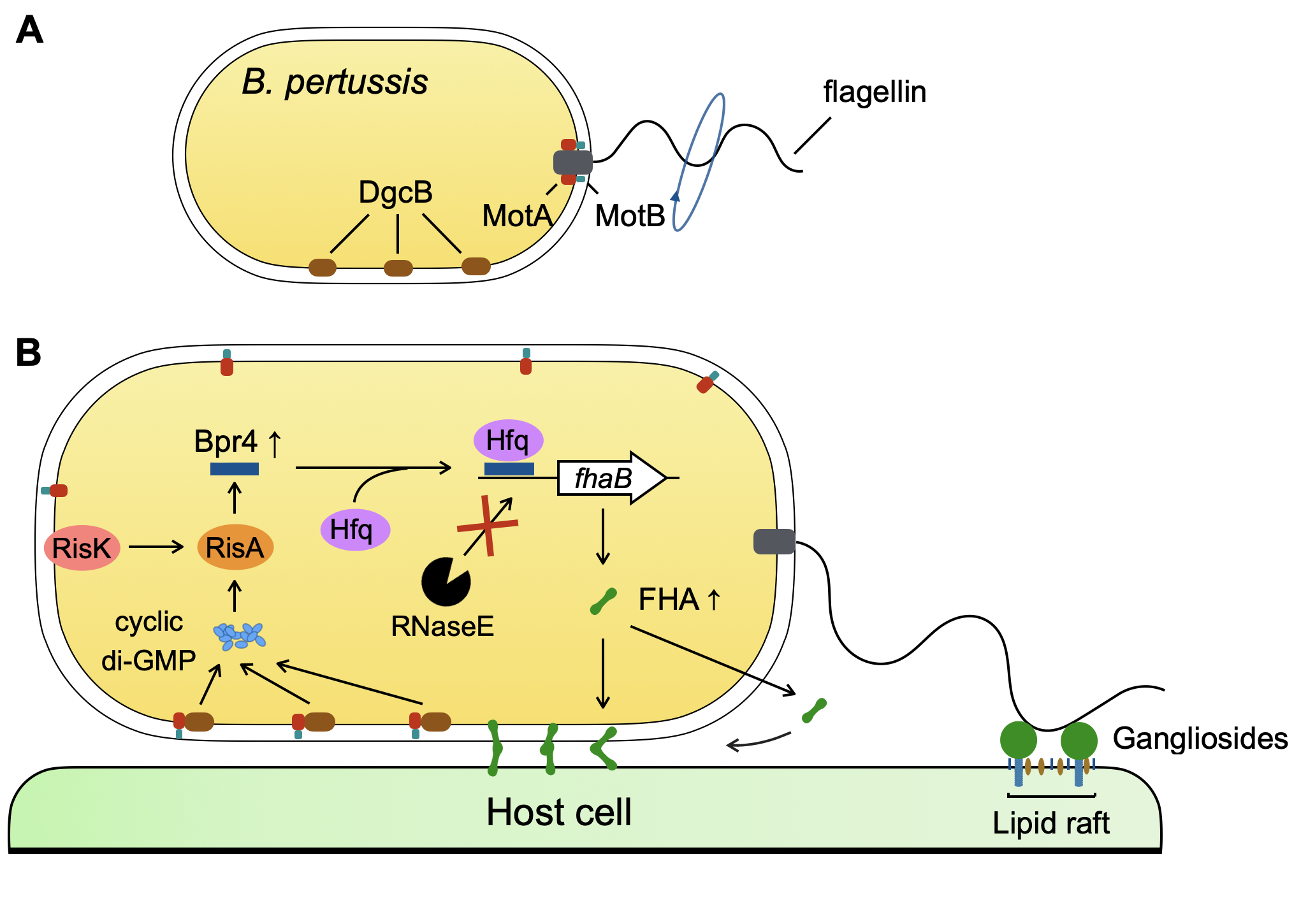
In the present study, we found that an sRNA, Bpr4, which is up-regulated up to 120-fold upon tracheal colonization of B. pertussis, binds to the 5’ untranslated region of fhaB mRNA encoding filamentous hemagglutinin (FHA), a major adhesin of B. pertussis, and protects it from RNaseE-mediated degradation, resulting in the posttranscriptional up-regulation of FHA, which facilitates bacterial colonization. Our results demonstrated that Bpr4 up-regulation is triggered by the interference of flagellar rotation after the interaction of flagellin and gangliosides on the host cells, which causes the disengagement of MotAB comprising a flagellar stator, from the flagellar complex. The liberated MotA interacted with and activated a diguanylate cyclase to generate cyclic di-GMP, which plays a role in Bpr4 up-regulation through the RisK/RisA two-component system. Our findings indicate that a flagellum-triggered sensory system leads to sRNA upregulation contributing to B. pertussis infection.
This article was published in Science Advances on Dec 22, 2022.
Title: “Interference of flagellar rotation up-regulates the expression of small RNA contributing to Bordetella pertussis infection”
Authors: Yukihiro Hiramatsu, Takashi Nishida, Dendi Krisna Nugraha, Mayuko Osada-Oka, Daisuke Nakane, Katsumi Imada, Yasuhiko Horiguchi
Links
-
Fig. The mechanism of sequential up-regulation of Bpr4 and FHA upon bacterial adherence to the host cell. (A) MotAB is incorporated into the flagellar basal body in motile B. pertussis. (B) Bpr4 is up-regulated upon cell adherence of B. pertussis through a flagellum-triggered sensory system. Hfq: RNA chaperon, DgcB: diguanylate cyclase B
- Home
- Achievement
- Research Activities
- Bordetella pertussis mechanosensing system contributing to bacterial colonization (Horiguchi Lab, in Sci. Adv.)