Genome-wide functional screening of drug-resistance genes in Plasmodium falciparum. (Iwanaga Lab, in Nat. Commun.)
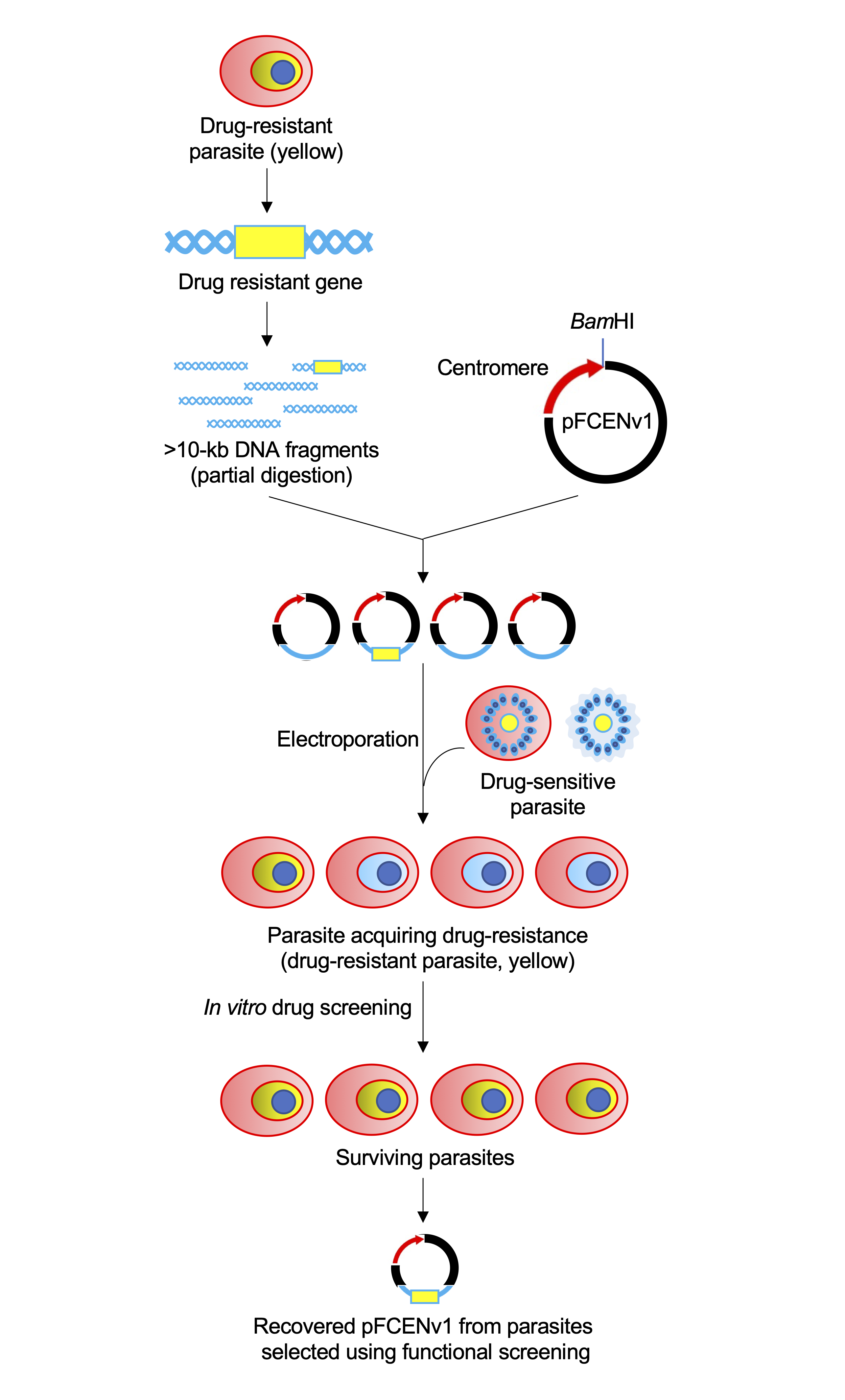
Prof. Iwanaga group developed the method for identifying drug resistance gene using Plasmodium artificial chromosome technology from P. falciparum. In addition, they succeeded to identify novel mefloquine resistance gene by using this developed method.
(ABSTRACT)
The global spread of drug resistance is a major obstacle to the treatment of Plasmodium falciparum malaria. The identification of drug-resistance genes is an essential step toward solving the problem of drug resistance. Here, we report functional screening as a new approach with which to identify drug-resistance genes in P. falciparum. Specifically, a high-coverage genomic library of a drug-resistant strain is directly generated in a drug-sensitive strain, and the resistance gene is then identified from this library using drug screening. In a pilot experiment using the strain Dd2, the known chloroquine-resistant gene pfcrt is identified using the developed approach, which proves our experimental concept. Furthermore, we identify multidrug-resistant transporter 7 (pfmdr7) as a novel candidate for a mefloquine-resistance gene from a field-isolated parasite; we suggest that its upregulation possibly confers the mefloquine resistance. These results show the usefulness of functional screening as means by which to identify drug-resistance genes.
This article was published in Nature communications on Oct 18, 2022.
Title: Genome-wide functional screening of drug-resistance genes in Plasmodium falciparum.
Journal: Nature communications
Authors:Shiroh Iwanaga, Rie Kubota, Tsubasa Nishi, Sumalee Kamchonwongpaisan, Somdet Srichairatanakool, Naoaki Shinzawa, Din Syafruddin, Masao Yuda, and Chairat Uthaipibull
DOI:10.1038/s41467-022-33804-w
Links
- Home
- Achievement
- Research Activities
- Genome-wide functional screening of drug-resistance genes in Plasmodium falciparum. (Iwanaga Lab, in Nat. Commun.)