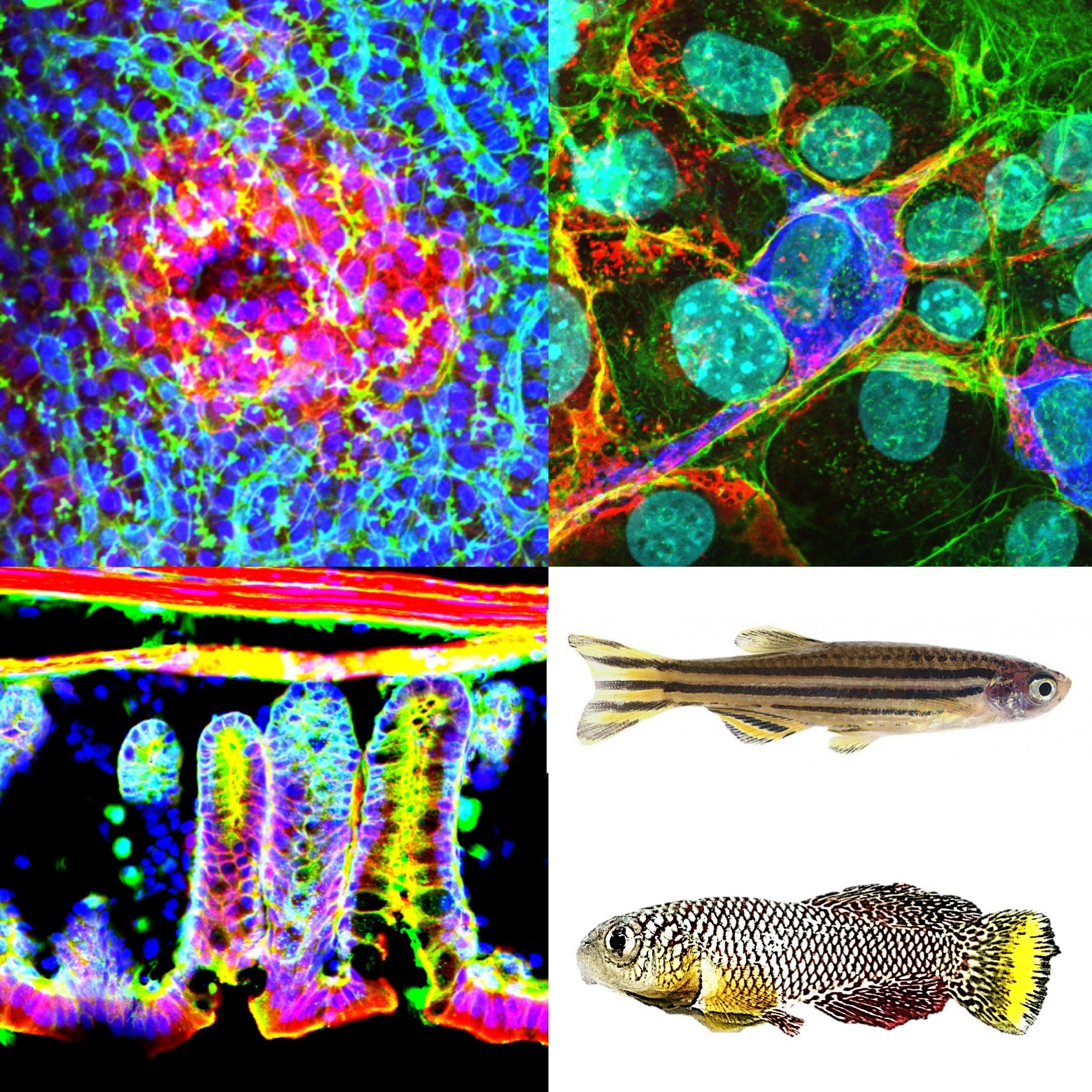
Ishitani Lab/Division of Cellular and Molecular Biology Department of Homeostatic Regulation
In our body, cells recognize their position and role and behave accordingly via cell-cell communication. Such behavior supports tissue morphogenesis and homeostasis, while its dysregulation is involved in congenital malformation, cancer, degenerative diseases, and aging. We focus especially on the cell-cell communication and behavior supporting tissue homeostasis and explore unknown molecular systems controlling embryonic development, organogenesis, regeneration, aging, and disease, using in vivo imaging, animal model genetics, molecular and cell biology, and biochemistry techniques.
"cell competition" a mechanisms ensuring developmental robustness and diseases prevention
Multicellular organisms, including humans, are constantly exposed to disturbances (noise). For example, replication errors occur during cell division, and reactive oxygen species produced by metabolism can damage the genome. Multicellular organisms are constantly exposed to such unavoidable noise, and by overcoming it in some way, they should achieve highly reproducible and robust development, regeneration, and homeostasis. We recently discovered that a phenomenon called "cell competition" is responsible for this noise removal (Nature Communications 2019, 2022, 2024; Science Advances 2024). Cell competition is a mechanism that senses and eliminates error cells (pathological cells) through intercellular communication. When this mechanism breaks down, pathological cells accumulate, cells are placed in inappropriate locations (such as spinal cord cells forming in the brain), motor functions do not develop properly, cancer occurs, and life spans are shortened. We are currently elucidating the mechanism of cell competition and analyzing its relationship to the onset of human diseases and cancer. We would like to use this knowledge to develop disease and cancer prevention technologies.
Prediction and control of human aging and lifespan
Another goal is to predict and control aging and lifespan. If we can suppress aging, we should be able to suppress the onset of all age-related diseases. In previous aging research, the time it takes to collect data (analyzing the aging process in humans takes decades, and even in mice it takes three years) has hindered progress. We have recently achieved aging analysis in a short period of time by using the ultra-short-lived fish killifish (lifespan 3-6 months) (Sci Rep 2022; NPJ Aging 2024; Science Advaces 2024). Furthermore, we are researching longevity factors from biodiversity data on people who live to be 100 or 110 years old or older, sharks that live for hundreds of years, and bats that live for decades, and introducing these factors into killifish to find substances that extend healthy lifespan. Furthermore, we are aiming to establish technology to predict remaining lifespan and future aging rates during health checkups.
Staff
- Prof.: Tohru Ishitani
- Asst. Prof.: Yuki Akieda
- Asst. Prof.: Kota Abe
- SA Asst. Prof.: Shizuka Ishitani
- SA Asst. Prof.: Kana Aoki
- JSPS Postdoc.: Shohei Ogamino
- JSPS Postdoc.: Kanako Matsumoto
Website
Publications
- (1) Foxo3-mediated physiological cell competition ensures robust tissue patterning throughout vertebrate development. Matsumoto K. and Akieda Y., et al., Nature Commun. (2024) 15:10662
(2) Mechano-gradients drive morphogen-noise correction to ensure robust patterning. Aoki K et al., Science Advances (2024) 10:eadp2357.
(3) Sex-dependent regulation of vertebrate somatic growth and aging by germ cells. Abe K et al., Science Advances (2024) 10:eadi1621
(4) Determining zebrafish dorsal organizer size by a negative feedback loop between canonical/non-canonical Wnts and Tlr4/NFκB. Zou J, et al., Nature Commun. (2023) 14:7194.
(5) Zebrafish imaging reveals additional TP53 mutation switching oncogene-induced senescence from suppressor to driver in primary tumorigenesis. Haraoka Y., et al., Nature Commun. (2022) 13:1417
(6) Intracellular pH controls WNT downstream of glycolysis in amniote embryos. Oginuma M., et al., Nature (2020) 584(7819):98-101.
(7) Cell competition corrects noisy Wnt/β-catenin morphogen gradients to achieve robust patterning in the zebrafish embryo. Akieda Y., et al., Nature Commun. (2019)10: 4710
(8) Hipk2 and PP1c cooperate to maintain Dvl protein levels required for Wnt signal transduction. Shimizu N., et al., Cell Reports (2014) 8(5) 1391-1404
(9) Visualization and exploration of Tcf/Lef function using a highly responsive Wnt/β-catenin signaling-reporter transgenic zebrafish. Shimizu N., et al., Developmental biology (2012) 370(1) 71-85
(10) Nemo-like kinase suppresses Notch signalling by interfering with formation of the Notch active transcriptional complex. Ishitani T., et al., Nature Cell Biol. (2010) 12:278-85
- Home
- Laboratories
- Ishitani Lab