Matsuura Lab/Research Center for Infectious Disease Control Laboratory of Virus Control
Our research is designed to elucidate the virus-host interactions involved in viral infection and pathogenicity. We develop therapeutic drugs and preventative programs for various viral infections based on our research results, with the aim of controlling infectious viruses in the human population.
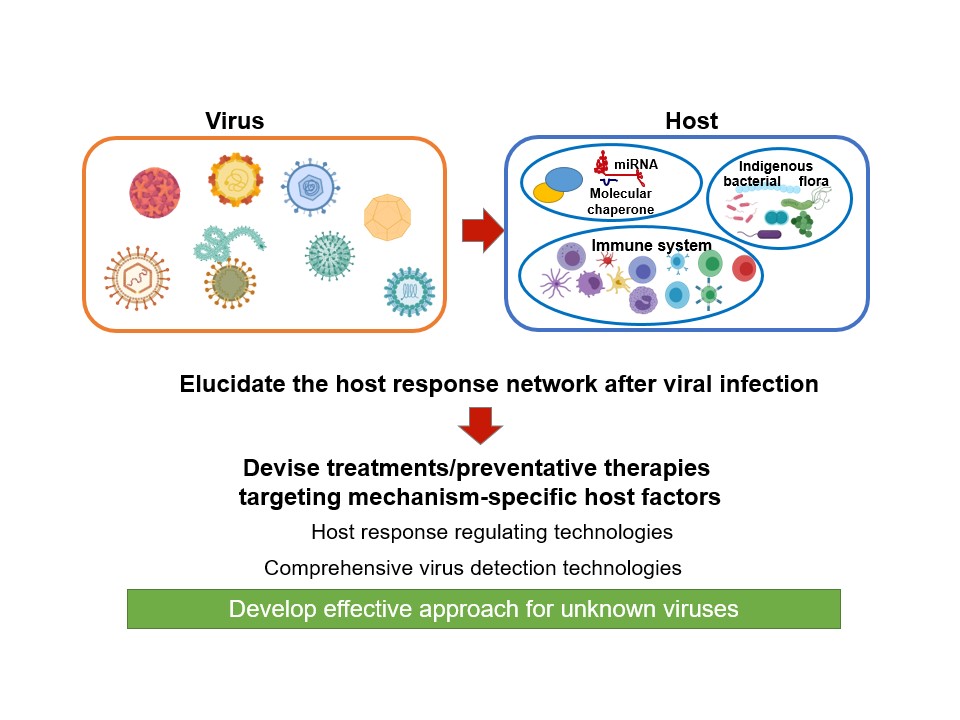
Analyzing the host-virus interaction network
With the changing climate and rapid expansion of human activity, modern society is plagued by countless emerging viral infections, which significantly damage both societies and economies. However, as revealed by the novel coronavirus (SARS-Cov-2) pandemic, it is difficult to predict outbreaks of emerging viral infections, and the development of control measures can be too late. We conduct research designed to develop a comprehensive understanding of the pathogenic mechanisms underlying viral infections using host-virus interactions in an attempt to enable the pre-emptive development of viral prophylactics and treatment strategies.
Specifically, we intend to create an animal model capable of reproducing human like viral pathogenesis in order to better characterize host responses to viral infection. This research is done in collaboration with researchers in immunology and molecular imaging allowing us to obtain more comprehensive data. This data will aid collaborations with researchers in data science and facilitate the construction of mathematical models of host response patterns. These data will enable the classification of viral infections by host response patterns, and facilitate of the development of new treatments targeting the host factors involved in the biological responses specific to each pattern.
Developing tools for viral research
To elucidate the pathogenesis of viruses and develop novel treatments and prevention strategies, researchers require tools that can quantify viral infectivity. We have developed a novel system for RNA virus research which includes the production of infective cDNA clones, virus-like particles, and pseudotype viruses to enable the study of highly virulent diseases without specialized BSL3 or 4 facilities. This system will be provided to the larger research community in the hope of accelerating viral research.
Staff
- SA Prof.: Yoshiharu Matsuura (concur.)
- SA Assoc.Prof.: Shuhei Taguwa (concur.)
- SA Assoc.Prof.: Chikako Ono (concur.)
- SA Assoc.Prof.: Saya Nakagomi (concur.)
- SA Assoc.Prof.:Ming-Te Yeh (concur.)
- SA Asst. Prof.: Junki Hirano
- SA Asst. Prof.: Kentaro Uemura (concur.)
- SA Asst. Prof.: Kazuma Okada (concur.)
- Postdoc. : Juan Vicente Bou Prados
Website
Publications
- (1)Establishment of a reverse genetics system for SARS-CoV-2 using circular polymerase extension reaction. Torii S., et al., Cell Reports (2021) 35(3):109014.
(2) Various miRNAs compensate the role of miR-122 on HCV replication. Ono C., et al., PLoS Pathog. (2020) 16(6):e1008308.
(3)Host ESCRT factors are recruited during chikungunya virus infection and are required for the intracellular viral replication cycle. Torii S., et al., J Biol Chem. (2020) 295(23):7941-7957.
(4) In vivo dynamics of reporter Flaviviridae viruses. Tamura T., et al., J Virol. (2019) 93(22):e01191-19.
(5) USP15 participates in HCV propagation through the regulation of viral RNA translation and lipid droplet formation. Kusakabe S., et al., J Virol. (2019) 93(6):e01708-18.
(6)Infection with flaviviruses requires BCLXL for cell survival. Suzuki T., et al., PLoS Pathog. (2018) 14(9):e1007299.
- Home
- Laboratories
- Matsuura Lab