Shioda Lab/Division of Infectious Disease Department of Viral Infections
Although we have been studying HIV for more than 20 years, now we are mainly studying mosquito-borne viral diseases such as dengue and chikungunya virus infections. We are conducting epidemiological studies in Thailand and molecular studies in Osaka, Japan. Recently, we are working on SARS-CoV-2.
- Molecular characterization of dengue and chikungunya viruses
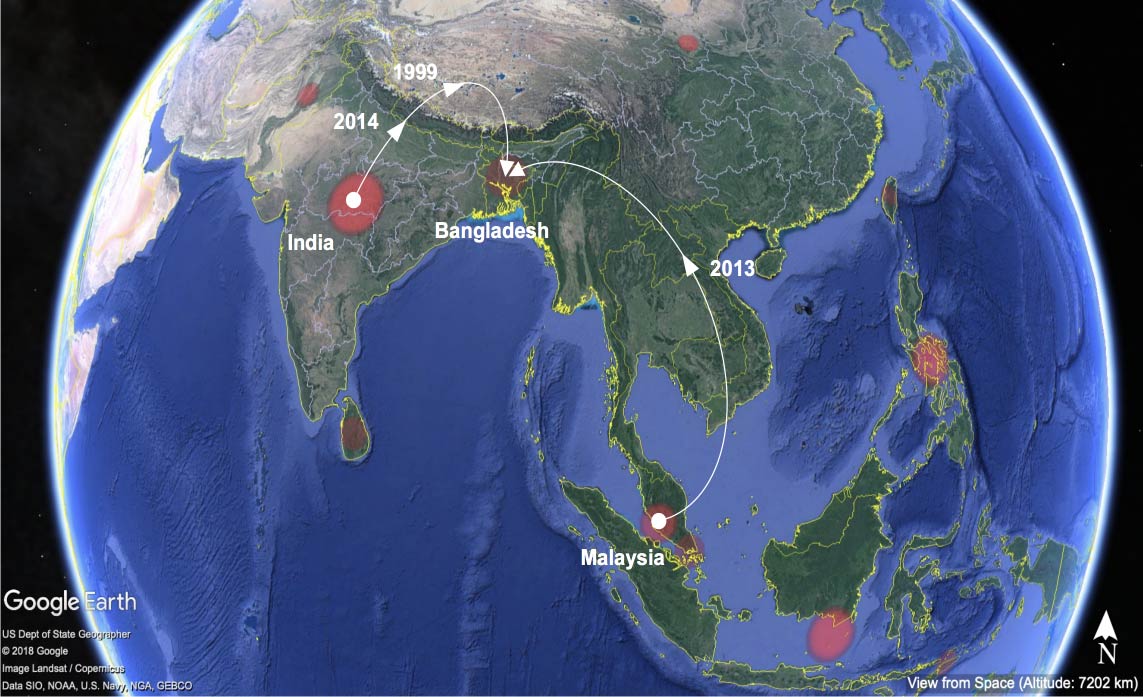
Dengue and chikungunya viruses are transmitted by Aedesmosquitos and cause febrile diseases. Dengue virus sometimes causes shock syndrome after the decline of fever and chikungunya virus causes arthralgia. We are conducting molecular epidemiology of these viruses in Thailand and Bangladesh by using molecular clock analysis. There are apparent variations in growth kinetics among isolated viruses and we are trying to elucidate factors affecting these differences.
- Characterization of anti-dengue antibodies
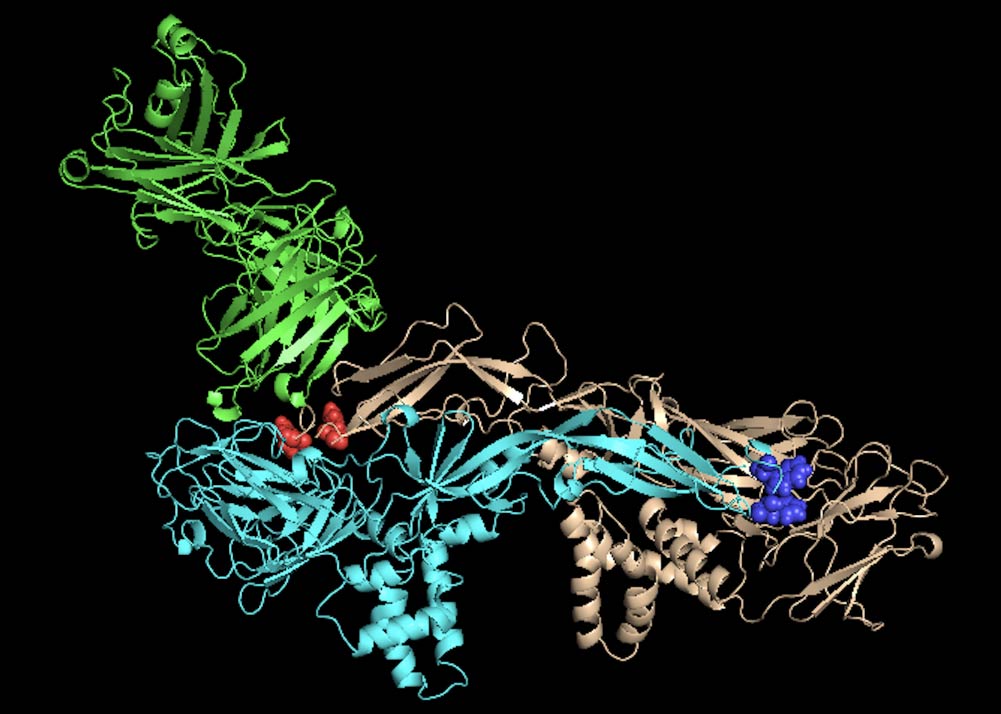
Anti-dengue antibodies show both neutralizing and enhancing effects on virus infection. We are analyzing several monoclonal antibodies hoping to find neutralizing antibodies without any enhancing effect. There are four serotypes of dengue virus. Some antibodies neutralize all four serotypes while others potently neutralize only one serotype. Antibodies with strong neutralizing activity without any enhancement can be used as antibody-drug. We will also analyze anti-viral antibodies in asymptomatic infection.
SARS-CoV-2
We are involved in the development of rapid diagnosis kits, study for the mechanisms of disease progression and phenotype of the isolated virus from patients’ specimens.
-
Fig. 1. Phylogeographical analysis of dengue virus type 2.
-
Fig. 2.A neutralizing antibody (green) and envelope dimer of dengue virus type 2 (pale blue and orange). Amino acid resides critical for antibody binding are highlighted with red and blue.
-
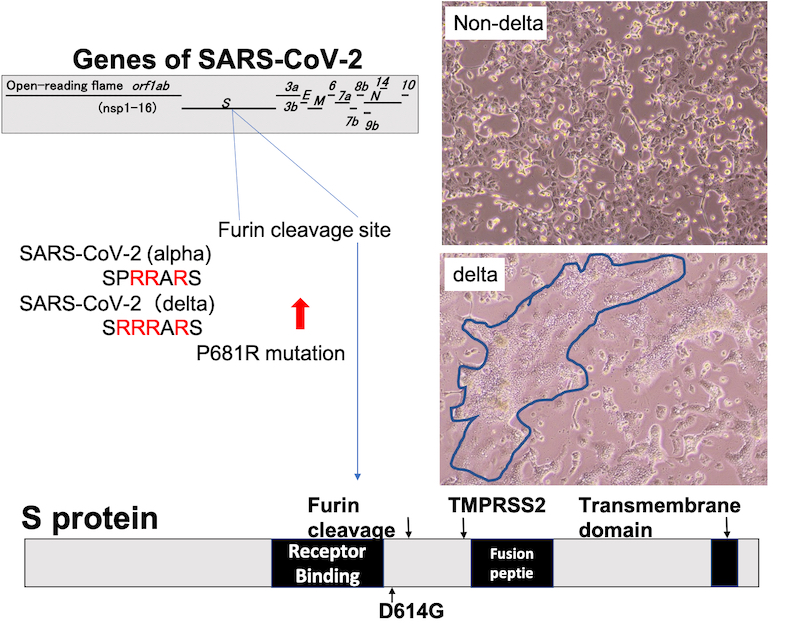
The schematic diagram of S protein with sequence of furin cleavage site. The cells surrounded by blue line indicate the big fusion caused by SARS-CoV-2 delta lineage infection.
Staff
- Prof.: Tatsuo Shioda
- Assoc. Prof.: Emi E. Nakayama
- Asst. Prof.: Tadahiro Sasaki
- Postdoc: Juthamas Phadungsombat
Website
Publications
- (1) Genetic regions affecting the replication and pathogenicity of dengue virus type 2. Samune Y, et al., PLoS Negl Trop Dis.(2024) 18:e0011885
(2) Enhancement of IL-6 Production Induced by SARS-CoV-2 Nucleocapsid Protein and Bangladeshi COVID-19 Patients' Sera. Hasan A, et al., Viruses. (2023) 15:2018.
(3) Changes in Anti-SARS-CoV-2 Antibody Titers of Pooled Plasma Derived From Donors in Japan: A Potential Tool for Mass-Immunity Evaluation. Yunoki M, et al., J Infect Dis. (2023) 228:889-894.
(4) Genetic Analysis of Dengue Virus in Severe and Non-Severe Cases in Dhaka, Bangladesh, in 2018-2022. Rahim R, et al., Viruses.(2023) 15:1144.
(5) COVID-19 relapse associated with SARS-CoV-2 evasion from CD4+ T-cell recognition in an agammaglobulinemia patient. Morita R, et al., iScience. (2023) 26:106685.
(6) Molecular Characterization of Dengue Virus Strains from the 2019-2020 Epidemic in Hanoi, Vietnam. Phadungsombat J,et al., Microorganisms. (2023) 11:1267.
(7) SARS-CoV-2 Related Antibody-Dependent Enhancement Phenomena In Vitro and In Vivo. Nakayama EE, et al., Microorganisms.(2023) 11:1015
(8) Anti-nucleocapsid antibodies enhance the production of IL-6 induced by SARS-CoV-2 N protein. Nakayama EE, et al., Sci Rep. (2022) 12:8108.
(9) Spread of a Novel Indian Ocean Lineage Carrying E1‐K211E/E2‐V264A of Chikungunya Virus East/Central/South African Genotype across the Indian Subcontinent, Southeast Asia, and Eastern Africa. Phadungsombat, J.et al., Microorganisms (2022) 10:354.
(10) Genetic Analysis of Influenza A/H1N1pdm Strains Isolated in Bangladesh in Early 2020. Hasan A, et al., Trop Med Infect Dis. (2022) 7:38.
- Home
- Laboratories
- Shioda Lab