/Research Collaboration Center for Infectious Diseases Section of Bacterial Drug Resistance Research
Carbapenem-resistant Enterobacteriaceae (CRE), including Klebsiella pneumoniae and Escherichia coli, are highly resistant to carbapenems and many other antibiotics. The rapidly increasing prevalence of CRE over the past decade has increased concern in healthcare facilities and public health communities worldwide. Japan is no exception, even though the prevalence of CRE at this time remains low. Our aim is to examine the epidemiological dissemination of CRE in the Southeast Asian countries. Carbapenem resistance is usually carried by plasmid(s) that harbor genes encoding carbapenemases, i.e., class A KPCs, class B metallo-β-lactamases (including IMP, VIM, or NDMs), or class D OXA-type enzymes. We have attempted to isolate CRE from patients admitted to leading hospitals in Thailand and Myanmar. CRE isolates are identified by biochemical characterization or MALDI-TOF-MS, followed by profiling using pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST). We then determine the whole genome sequence of CRE isolates to identify the full plasmid and construct a comprehensive image of the relationships between isolates based on MLST and phylogeny. By undertaking these genomic epidemiological studies, we increase our understanding of how CRE spread and may be able to identify potential reservoirs.
-
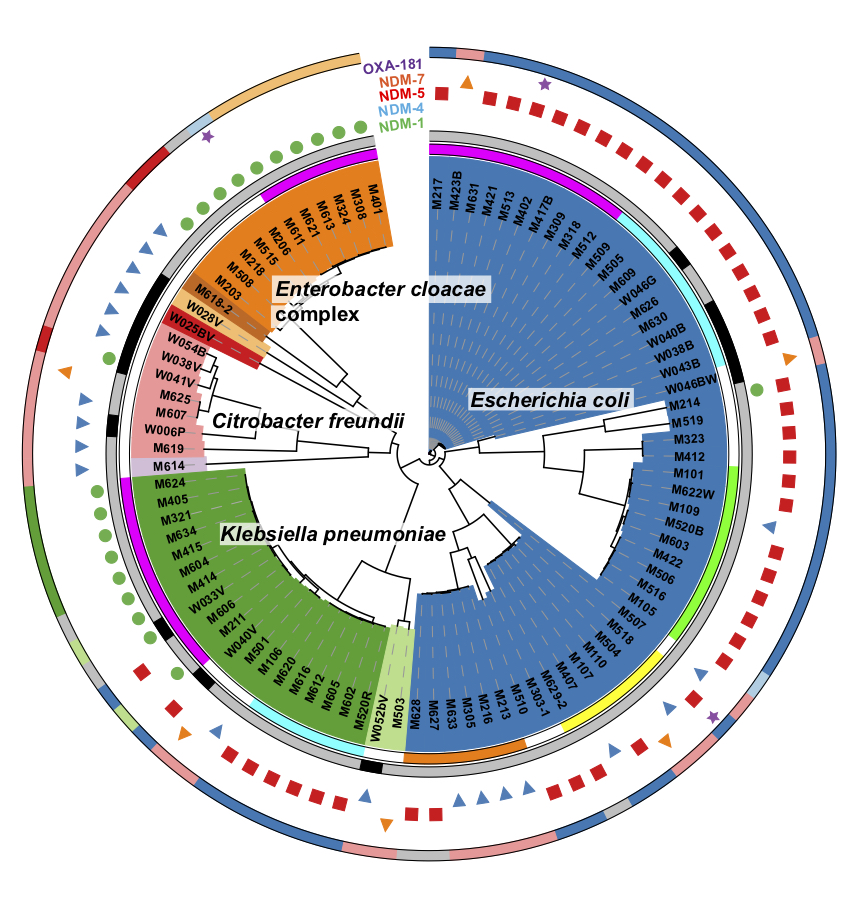
A whole genome SNP-based phylogenetic tree of CRE isolates from Myanmar. The inner colored regions define bacterial species. Next outer colored regions denote the origins of isolates. Colors and sizes of outer dots denote classes and numbers of antimicrobial resistance genes harbored by each isolate.
Staff
- Prof.: Tetsuya Iida (concur.)
- Assoc. Prof.: Shigeaki Matsuda (concur.)
- SA Assoc. Prof.:Ryuichiro Abe(concur.)
Website
Publications
- (1) In Virro Efficacy of Meropenem-Cefmetazole Combination Therapy against New Delhi Melallo-β lactamase-producing Enterobacteriaceae. Hagiya H, et. al. lnt J Antimicrob Agents. (2020) 55:105905.
(2) Genomic characterization or an emerging blaKPC-2 carrying Enterobacteriaceae clinical isolates in Thailand. Kerdsin A. el al. Sci Rep. (2019) 9:18571
(3) Genomic characterisation of a novel plasmid carrying blaIMP-6 of carbapenem-resistant Klebsielta pnevmoniae isolated in Osaka, Japan. Abe R et al. J Glob Anlimicrob Resist. (2019) pii: S2213·7165 (19)30257-7.
(4) Dissemination of carbapenemase-producing Entnrobacteriacese harbouring blaNDM or blaIMI in local market foods of Yangon, Myanmar. Sugawara Y. et. al Sci Rep. (2019) 9:14455.
(5) Rapid screening and early precautions for carbapenem-resistant Acinetobacter baumannii carriers decreased nosocomial transmission in hospital settings: a quasi-experimental study. Yamamoto N. et. al. Antimicrob Resist Infect Control.(2019) 8:110