/Research Collaboration Center for Infectious Diseases Section of Bacterial Infections
In Thailand, enteric infections caused by bacteria, viruses, and protozoa are a regular occurrence, posing a significant public health threat. In recent years, emerging and reemerging infectious diseases have also become major concerns, amplifying the burden on the public health system. These infections have the potential to spread rapidly, increasing health risks across society.
Rapid pathogen detection and infectious disease monitoring
To address these challenges, it is essential to strengthen systems for rapidly detecting pathogens and continuously monitoring infectious disease outbreaks. In collaboration with the Thai Ministry of Public Health’s Department of Medical Sciences and the Epidemiology Division of the Department of Disease Control, our section is performing research targeting patients with severe diarrheal diseases, including cholera.
Our primary research objectives are as follows:
・Early detection of causative pathogens
・Development of diagnostic methods and preventive measures
To achieve these goals, we employ the latest molecular biology techniques to improve traditional pathogen detection methods and facilitate rapid responses to emerging and reemerging infectious diseases. We use bacterial culture methods, real-time PCR, next-generation sequencing, and time-of-flight mass spectrometry (TOF-MS) for the early detection of pathogens, enabling faster and more accurate identification and enhancing our capacity to respond effectively to infectious threats.
Response to infectious diseases based on epidemiological investigations
We perform epidemiological investigations of outbreaks and genomic analysis of the causative microorganisms to identify the infection sources, transmission routes, and key pathogens. These efforts enable the early detection of high-risk or previously unknown pathogens and deepen our understanding of the factors contributing to the onset of diarrhea and group diarrhea outbreaks. We aim to use our findings to establish effective intervention strategies to prevent future outbreaks.
The results of our research are intended to contribute to disease control efforts within Thailand and strengthen outbreak response capabilities in neighboring Asian countries and Japan.
-
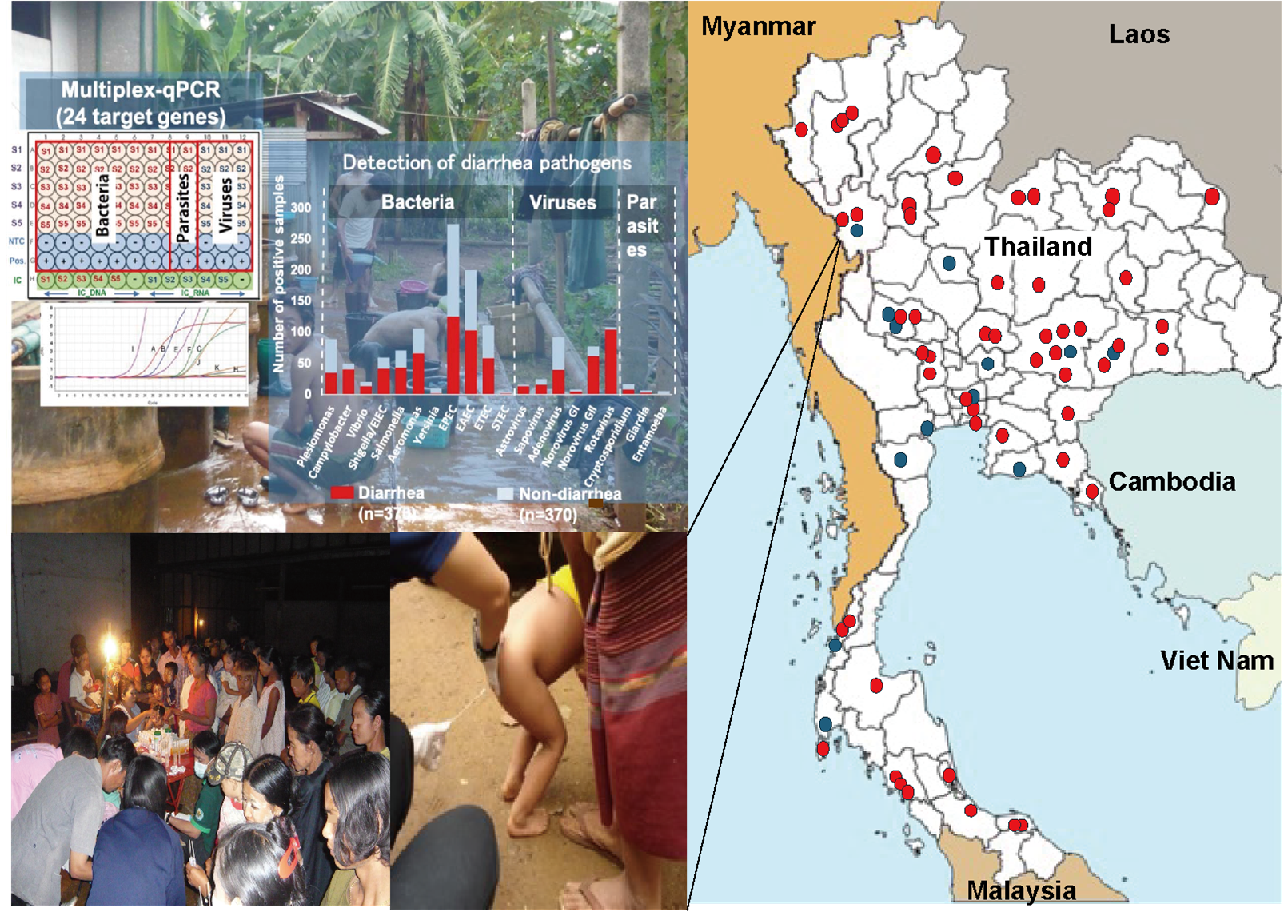
Fig. Multiple outbreaks (●) and sporadic cases (●) have been investigated in collaboration with the Department of Disease Control and the Department of Medical Sciences, Ministry of Public Health, Thailand. Investigations of Vibrio cholerae and other enteropathogens in border areas (lower left photo) were conducted. Detection and analysis of pathogens are performed using multiplex real-time PCR methods and genome sequencing
Staff
- Prof.: Tetsuya Iida (concur.)
- SA Assoc.Prof.: Kazuhisa Okada
Publications
(1) First recorded food-borne outbreak of gastroenteritis caused by enteroinvasive Escherichia coli serotype O8:H19 in Thailand. Okada K. et al., Eur J Clin Microbiol Infect Dis (2025) 44:733-737.
(2) Flagella-related gene mutations in Vibrio cholerae during extended cultivation in nutrient-limited media impair cell motility and prolong culturability. Okada K. et al., mSystems (2023) e0010923.
(3) Emergence of Vibrio parahaemolyticus serotype O10:K4 in Thailand. Okada K. et al., Microbiol Immunol. (2023) 67: 201-203.
(4) Etiologic features of diarrheagenic microbes in stool specimens from patients with acute diarrhea in Thailand. Okada K. et al., Sci. Rep. (2020) 10:4009.
(5) Simultaneous detection and quantification of 19 diarrhea-related pathogens with a quantitative real-time PCR panel assay. Wongboot W. et al., J Microbiol Methods. (2018) 151:76-82
(6) Vibrio cholerae embraces two major evolutionary traits as revealed by targeted gene sequencing. Okada K., et al., Sci. Rep. (2018) 8(1):1631
(7) Characterization of 3 Megabase-Sized Circular Replicons from Vibrio cholerae. Okada K., et al., Emerg Infect Dis. (2015) 21(7):1262-3.
(8) Cholera in Yangon, Myanmar, 2012-2013. Aung WW., et al., Emerg Infect Dis. (2015) 21(3):543-4.
(9) Vibrio cholerae O1 isolate with novel genetic background, Thailand–Myanmar. Okada K., et al., Emerg Infect Dis. (2013) 19:1015-7.