Iida Lab/Research Center for Infectious Disease Control Department of Bacterial Infections
In our laboratory, we are conducting research and collecting genomic information to understand how bacterial pathogens infect the host and cause disease. In addition, by developing new pathogen detection methods using high-throughput DNA sequencers, we aim to identify novel pathogens and reveal the pathogenesis of unknown infectious diseases.
Identifying the mechanism(s) underlying bacterial infection and pathogenesis
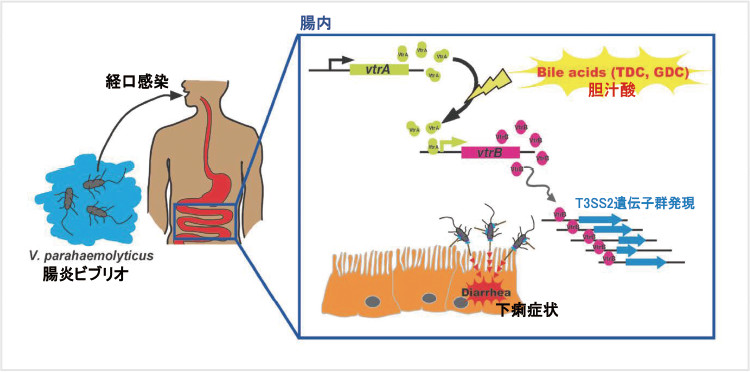
We performed whole genome sequencing of Vibrio parahaemolyticus, a bacterium that causes acute gastroenteritis in humans, and revealed that the type III secretion system T3SS2 is essential for pathogenicity. T3SS2 directly injects bacterial proteins (effectors) into target host cells. We demonstrated that injection of those effectors by T3SS2 from V. parahaemolyticus leads to inflammation of the intestinal mucosa and diarrhea. Currently, we are analyzing the molecular mechanism by which those effectors cause the symptoms of acute gastroenteritis.
Also, we revealed that expression of the genes encoding T3SS2 is induced by bile. In fact, chemical substances that adsorb and remove bile suppressed symptoms caused by V. parahaemolyticus in animal models, suggesting that these substances may be new therapeutic agents for V. parahaemolyticus infection. This is an example of “anti-virulence therapy” rather than antimicrobial therapy. This kind of approach is expected to provide novel therapeutic strategies for various bacterial infections.
Furthermore, based on findings obtained from our research on pathogenicity, we aim to explore the life cycle of bacterial pathogens in their natural environments.
Development of methods to diagnose bacterial infections based on genomics and metagenomics
Emerging and re-emerging infectious diseases cause many problems worldwide. In many cases of such infection, the causative agent is unknown and/or the pathogenic mechanism is not yet clear. To identify the agents that cause such infections, and to understand the underlying pathogenesis, we are developing a high-throughput DNA sequencing-based system to detect pathogens and analyze their virulence traits.
Staff
- Prof.: Tetsuya Iida
- Assoc. Prof.: Shigeaki Matsuda
- Asst. Prof.: Eiji Ishii
- Sa Asst. Prof.: Andre Pratama
- Postdoc.: Shymaa Saeed Ali
Website
Publications
- (1) The read-through transcription-mediated autoactivation circuit for virulence regulator expression drives robust type III secretion system 2 expression in Vibrio parahaemolyticus. Anggramukti DS. et al., PLoS Pathog. (2024) 10.1371/journal.ppat.1012094
(2)The Xenogeneic Silencer Histone-Like Nucleoid-Structuring Protein Mediates the Temperature and Salinity-Dependent Regulation of the Type III Secretion System 2 in Vibrio parahaemolyticus. Pratama A. et al., J Bacteriol. (2022) 10.1128/jb.00266-22
(3) Export of a Vibrio parahaemolyticus toxin by the Sec and type III secretion machineries in tandem. Matsuda S., et al., Nat. Microbiol. (2019) 4:781-8.
(4) A repeat unit of Vibrio diarrheal T3S effector subverts cytoskeletal actin homeostasis via binding to interstrand region of actin filaments. Nishimura M., et al., Sci Rep. (2015) 5:10870.
(5) Interaction between the type III effector VopO and GEF-H1 activates the RhoA-ROCK pathway. Hiyoshi H., et al., PLoS Pathog. (2015) 11(3):e1004694.
(6) A cytotoxic type III secretion effector of Vibrio parahaemolyticus targets vacuolar H+-ATPase subunit c and ruptures host cell lysosomes. Matsuda S., et al., PLoS Pathog. (2012);8(7):e1002803.
(7) VopV, an F-actin-binding type III secretion effector, is required for Vibrio parahaemolyticus-induced enterotoxicity. Hiyoshi H., et al., Cell Host Microbe. (2011) 10(4):401-9. doi: 10.1016/j.chom.2011.08.014.
(8) Metagenomic diagnosis of bacterial infections. Nakamura S., et al., Emerg Infect Dis. (2008) 14(11):1784-6.
- Home
- Laboratories
- Iida Lab