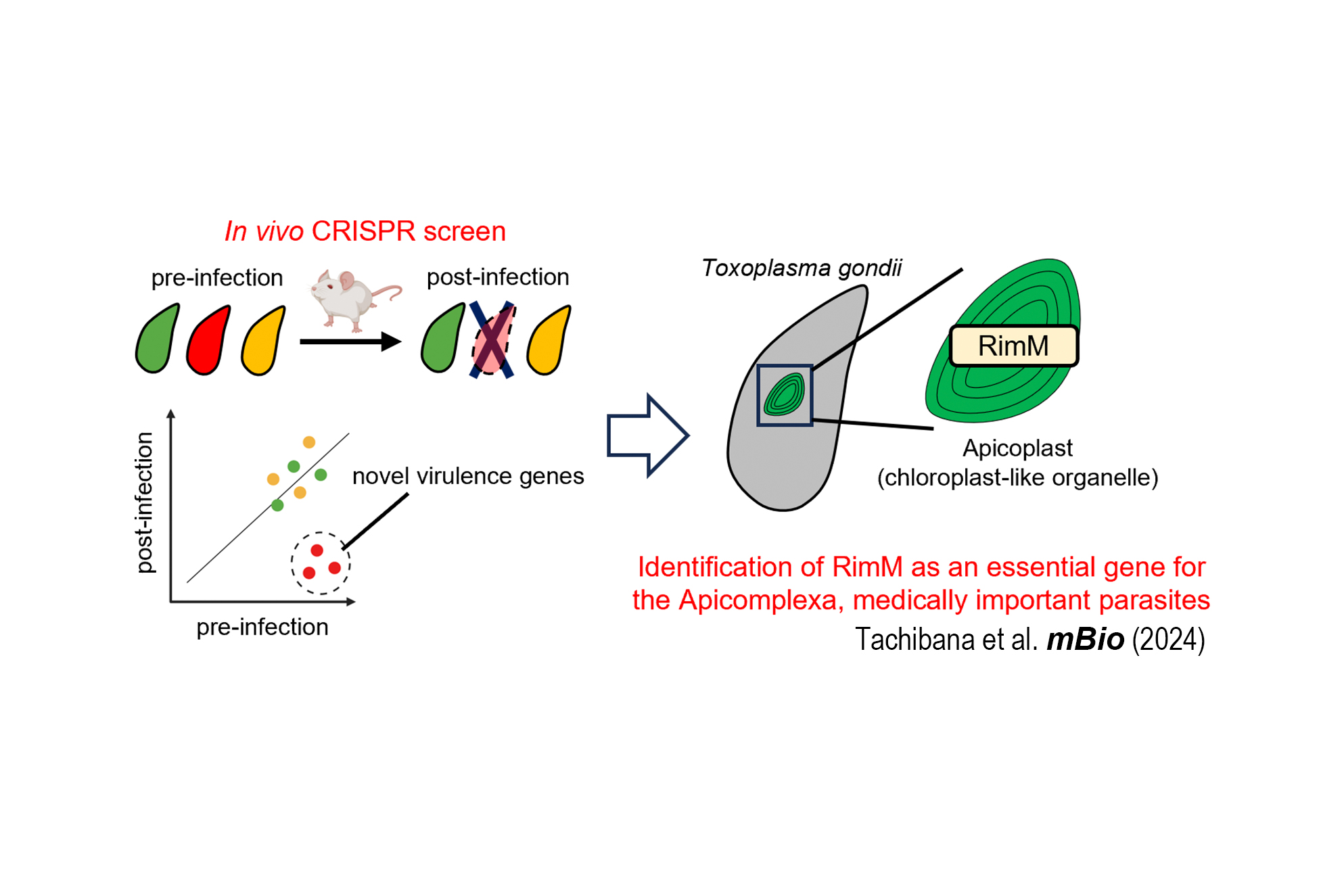
CRISPR screens identify genes essential for in vivo virulence among proteins of hyperLOPIT-unassigned subcellular localization in Toxoplasma (Yamamoto Lab, in mBio)
The research field to identify and characterize genes essential for in vivo virulence in Toxoplasma gondii has been dramatically advanced by a series of in vivo clustered regularly interspaced short palindromic repeats (CRISPR) screens. Although subcellular localizations of thousands of proteins were predicted by the spatial proteomic method called hyperLOPIT, those of more than 1,000 proteins remained unassigned, and their essentiality in virulence was also unknown. In this study, we generated two small-scale gRNA libraries targeting approximately 600 hyperLOPIT-unassigned proteins and performed in vivo CRISPR screens. As a result, we identified several genes essential for in vivo virulence that were previously unreported. We further characterized two candidates, TgGTPase and TgRimM, which are localized in the cytoplasm and the apicoplast, respectively. Both genes are essential for parasite virulence and widely conserved in the phylum Apicomplexa. Collectively, our current study provides a resource for estimating the in vivo essentiality of Toxoplasma proteins with previously unknown localizations.
This article was published in mBio on July 31, 2024
Title: CRISPR screens identify genes essential for in vivo virulence among proteins of hyperLOPIT-unassigned subcellular localization in Toxoplasma
Authors: Yuta Tachibana, Miwa Sasai and Masahiro Yamamoto
Links
- Home
- Achievement
- Research Activities
- CRISPR screens identify genes essential for in vivo virulence among proteins of hyperLOPIT-unassigned subcellular localization in Toxoplasma (Yamamoto Lab, in mBio)