Evolutionary dynamics of heparan sulfate utilization by SARS-CoV-2 (Arase Lab, in mBio)
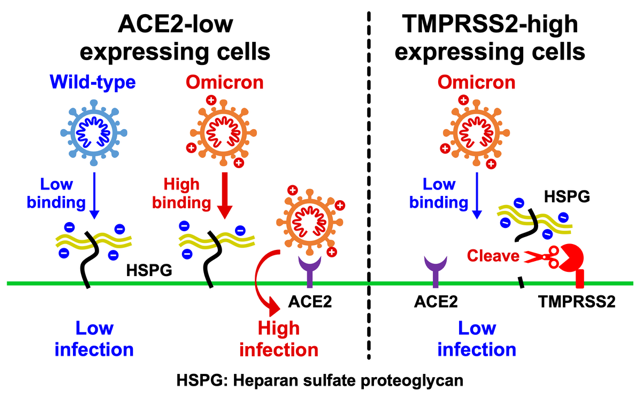
The SARS-CoV-2 Omicron variant acquired high infectivity through more than 30 mutations in its spike protein, but the detailed molecular mechanism remained unclear. This study reveals that mutations in the Omicron spike protein increased the number of positively charged amino acid residues, thereby enhancing its affinity for negatively charged heparan sulfate on the cell surface. We suggest that this enhanced affinity enables efficient infection of cells with low ACE2 receptor expression.
On the other hand, while conventional strains exhibit high infectivity in cells expressing the TMPRSS2 protease, such as alveolar epithelial cells, the Omicron variant shows reduced infectivity in these cells. Our research newly discovered that heparan sulfate proteoglycans on the cell surface are cleaved by TMPRSS2, which may account for this phenomenon.
These findings suggest that by acquiring the ability to bind to heparan sulfate, the Omicron variant not only enhanced its capacity to infect cells with low ACE2 expression but also altered its overall viral cell tropism. The results of this study provide critical insights into understanding how mutations in the SARS-CoV-2 spike protein affect the virus's infectivity, cell tropism, and, consequently, its pathogenicity.
Audio Guide to this Research
This article was publishet in mBio, on June 23, 2025
Title: Evolutionary dynamics of heparan sulfate utilization by SARS-CoV-2
Authors: Shuhei Higuchi Yafei Liu, Jun Shimizu, Chikako Ono, Yumi Itoh, Wataru Nakai, Hui Jin, Kazuki Kishida, Kazuo Takayama, Toru Okamoto, Yoshiko Murakami, Taroh Kinoshita, Yoshiharu Matsuura, Tatsuo Shioda, Hisashi Arase
Links
- Home
- Achievement
- Research Activities
- Evolutionary dynamics of heparan sulfate utilization by SARS-CoV-2 (Arase Lab, in mBio)