Yamasaki Lab/Bioinformatics Center Department of Biological Informatics Laboratory of RNA Informatics
The RNA Informatics Laboratory aims to elucidate the mechanism underlying gene expression regulation by analyzing messenger RNA (mRNA). Subsequently, applying this knowledge will remarkably contribute to society.
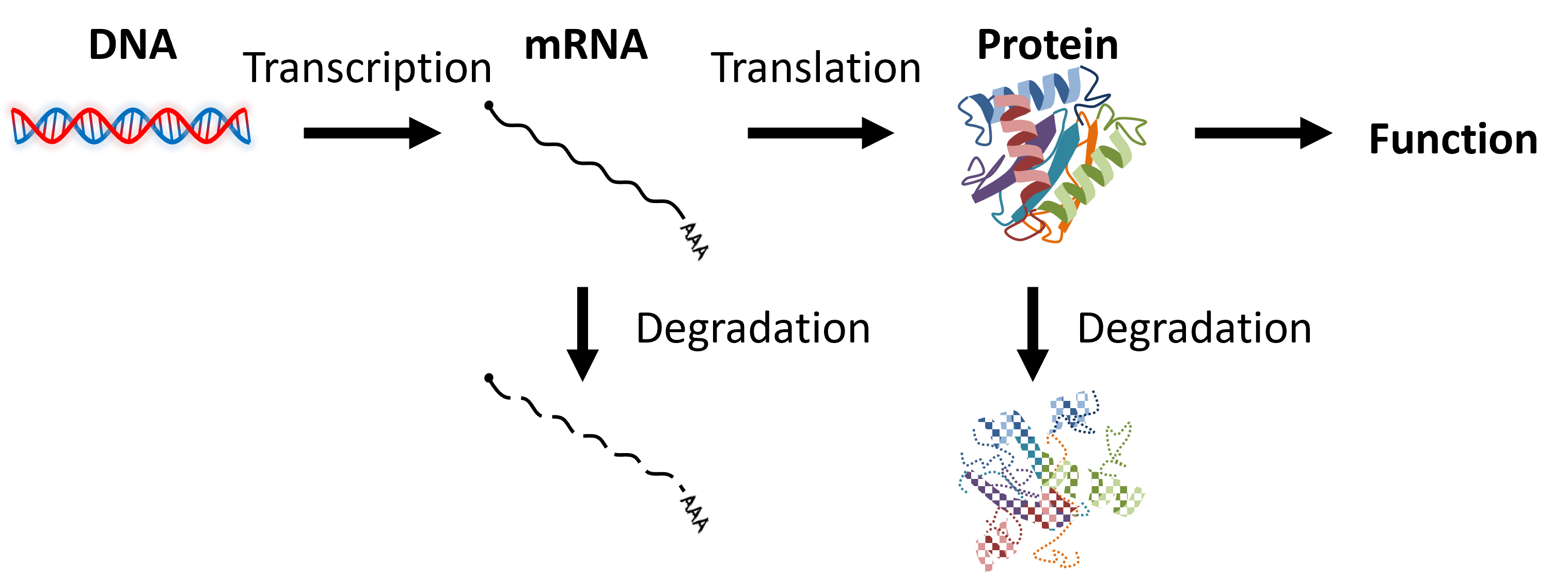
Various proteins in the human body, including enzymes and structural proteins, play essential roles in sustaining life. Although the genetic blueprint for these proteins is encoded in DNA, the information in DNA is not directly converted into proteins. Instead, it is initially transcribed into mRNA and then translated into proteins (Figure 1). This series of processes is known as gene expression, and the timing and quantity of protein production must be precisely regulated in accordance with the cell type and environmental conditions for life to function properly.
The final level of protein expression is determined by mRNA transcription efficiency, mRNA stability, mRNA translation efficiency, and protein stability. However, disruptions in these regulatory mechanisms, whether caused by genetic or environmental factors, can lead to various diseases, including genetic disorders and cancer. Therefore, comprehensively understanding gene expression regulation is crucial for uncovering fundamental biological principles and the causes of diseases, and mRNA plays a pivotal role in this understanding.
Furthermore, mRNA research and its associated technologies are vital for drug development and biotechnology advancements. For example, mRNA-based vaccines and gene therapies provide innovative treatment strategies that are different from traditional pharmaceuticals. In addition, in the large-scale production of therapeutic and industrial proteins, mRNA serves as a crucial blueprint. Optimizing mRNA sequences can enhance biopharmaceutical efficiency and enzyme production.
Given the abovementioned factors, mRNA research is essential for the advancement not only in life sciences but also in the medical and biotechnology industries. Our laboratory uses advanced sequencing technologies and machine learning to analyze the complex regulatory mechanisms of mRNA transcription, degradation, and translation. The findings of our research will provide insights into the development of new medical and industrial technologies.
Understanding and Applying the Mechanisms of mRNA Sequence Determination
Transcription is the first step in gene expression, which influences all subsequent processes. During this phase, the amount of mRNA transcribed is precisely regulated in accordance with the cell type and environmental conditions. Equally important is the mechanism by which the specific sequence of mRNA is determined, indicating the transcribed mRNA variants.
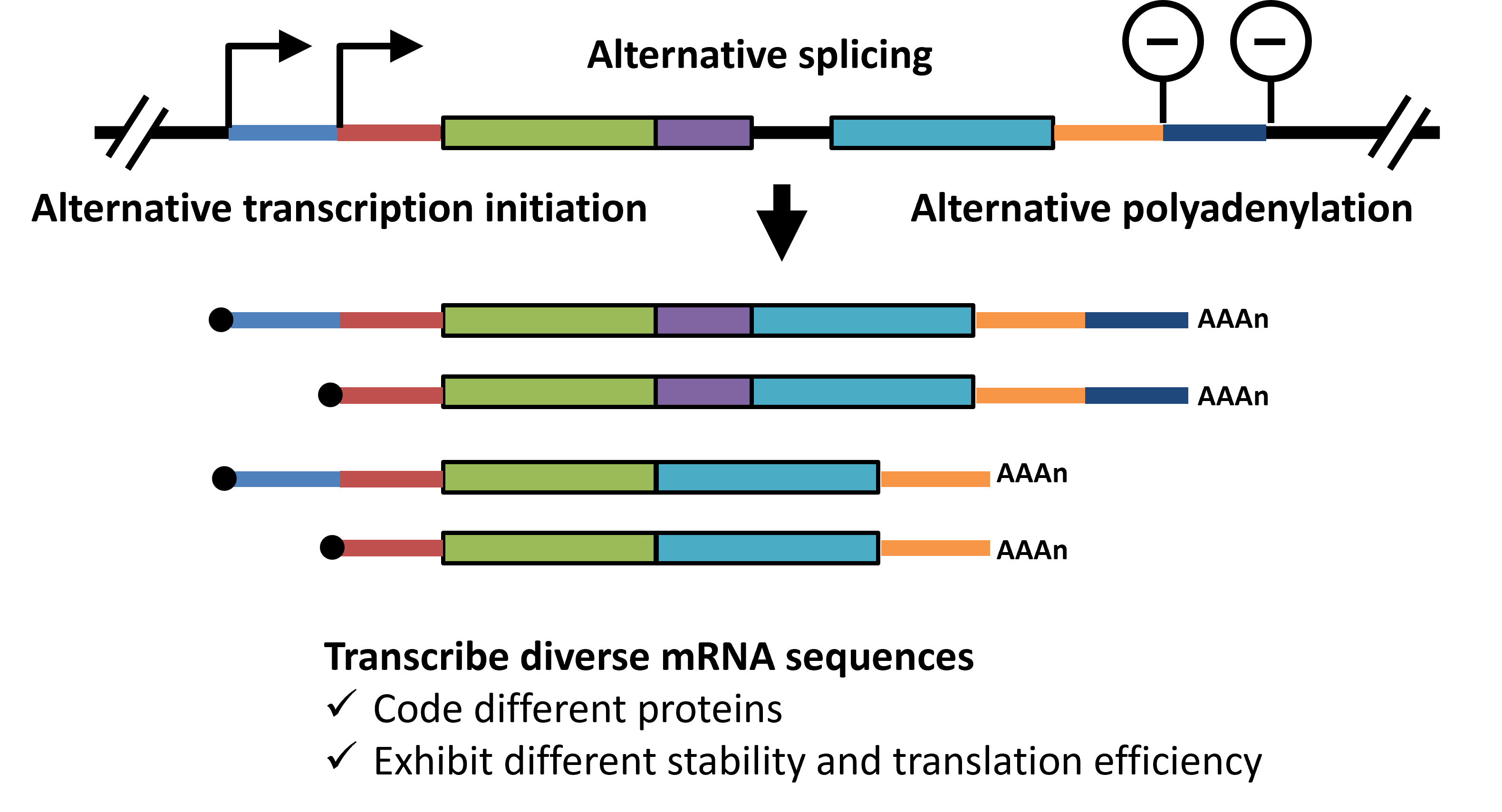
The number of protein-coding genes in humans is approximately 20,000, which is not significantly different from the 18,000 genes found in Chlamydomonas reinhardtii (a unicellular green alga). However, humans have evolved highly advanced post-transcriptional regulatory mechanisms, such as alternative splicing, which allows a single gene to produce multiple mRNA isoforms, thereby dramatically increasing the diversity of proteins (Figure 2). Furthermore, alternative transcription initiation and polyadenylation produce multiple mRNA isoforms that encode the same protein. However, their stability and translation efficiency differ from one another. This diversity allows for a highly complex gene expression regulation, which is essential for complex biological functions. Disruption of these selective transcription mechanisms has been linked to the onset and progression of various diseases.
Thus, our laboratory analyzes the mechanism that determine transcription start sites, splicing patterns, and polyadenylation sites. Using the insights obtain from analysis, we develop a predictive system to model and understand selective transcription mechanisms. In this research, we aim to detect DNA mutations that affect selective transcription mechanisms and optimize gene sequences for the efficient production of valuable proteins.
Understanding and Applying the Mechanisms of mRNA Stability and Translation Efficiency
Similar to transcription efficiency, mRNA stability and translation efficiency are crucial regulatory stages of gene expression. For example, transcription factors must respond rapidly to environmental changes and signaling pathways; therefore, the mRNAs encoding them are unstable and quickly degraded when no longer needed. By contrast, structural proteins, which require stable and high levels of expression, are encoded by more stable mRNAs. Translation efficiency is often correlated with protein accumulation levels, making it the second most crucial determinant of gene expression after transcription. Moreover, mRNA translation efficiency plays a role in regulating cellular energy consumption and in adjusting the translation efficiency of various biosynthetic genes in accordance with cellular conditions.
The stability and translation efficiency of mRNA are determined by diverse regulatory sequences located in the 5′ untranslated region, coding sequence, and 3′ untranslated region. Our laboratory integrates transcription inhibition chase assays and polysome fractionation with CAGE-seq and nanopore long-read cDNA sequencing, allowing for more precise and comprehensive analysis of mRNA isoforms. In this research, we aim to elucidate the key factors involved in the regulation of mRNA stability and translation efficiency by combining this high-precision data with comprehensive feature analysis, statistical methods, and machine learning approaches. The findings of this research are further integrated with the data on selective transcription mechanisms, enabling us to advance research on gene expression regulation across the stages of transcription, degradation, and translation.
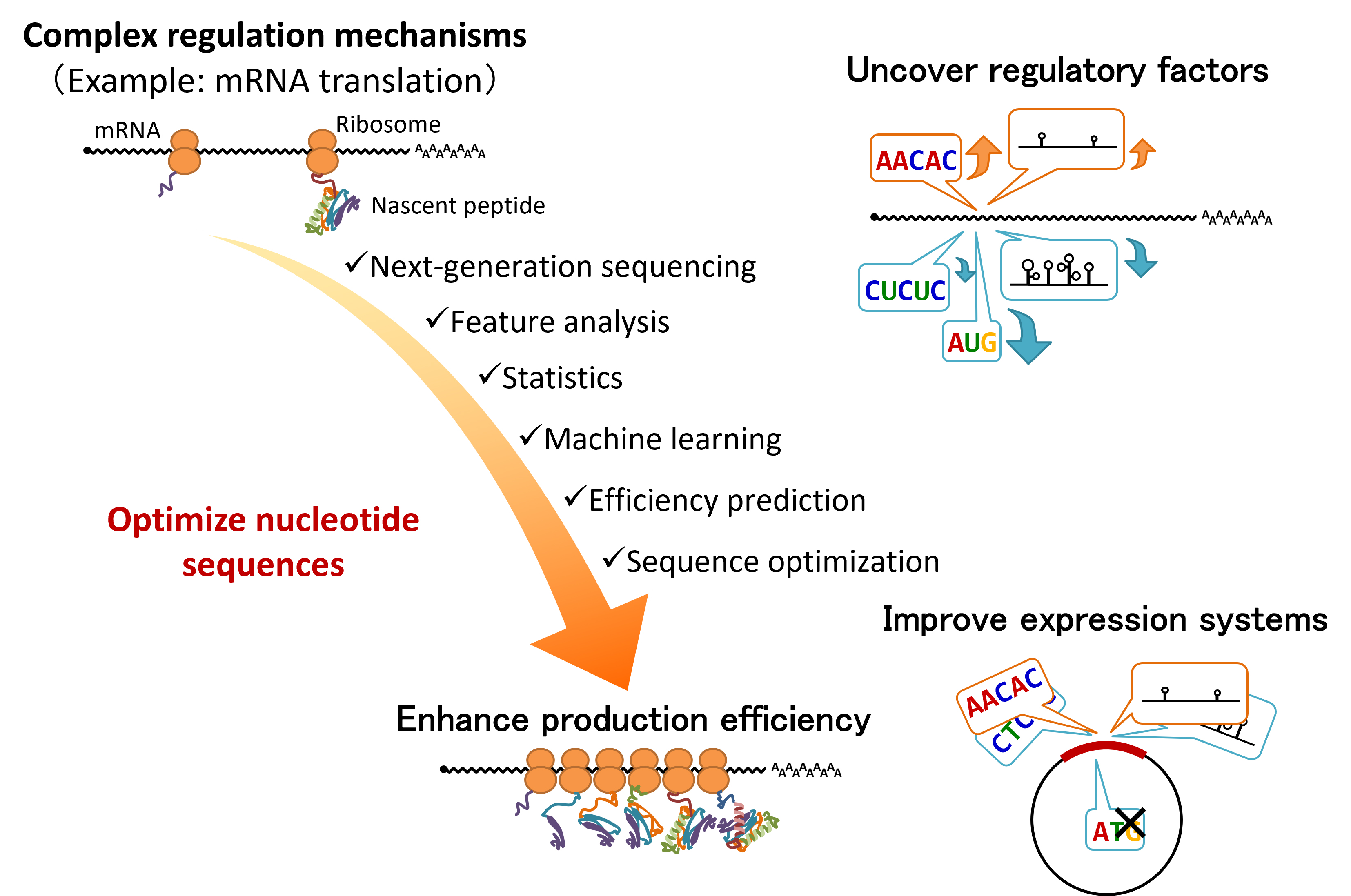
Furthermore, mRNA stability and translation efficiency are crucial in the development of mRNA therapeutics and in the large-scale production of valuable proteins. In the medical and biotechnology industries, highly stable and efficiently translated mRNA sequences are necessary. Thus, our laboratory is developing an mRNA sequence optimization system (Figure 3) by utilizing the collected data, which are large-scale datasets obtained from massively parallel reporter assays, machine learning, and genetic algorithms. This system incorporates a highly accurate prediction model based on high-quality data, allowing for sustained and efficient protein synthesis compared with conventional methods.
Promoting the Applications of RNA
The applications of RNA are rapidly expanding across various fields, including medicine, agriculture, livestock farming, aquaculture, and the biotechnology industry. Thus, our laboratory is actively developing RNA sequence optimization systems to improve RNA stability and translation efficiency, thereby contributing to the diverse applications of RNA in these fields.
For example, mRNA sequences are optimized for cell-free protein synthesis. Cell-free protein synthesis is a technique that enables protein production using extracted cellular components without the need for living cells. A key advantage of this method is its ability to rapidly synthesize proteins without the need for cell culture. Considering that mRNA can be directly used as a template, genetic modification is not necessary.
In our laboratory, massively parallel reporter assays are used to obtain detailed data on mRNA stability and translation efficiency in cell-free protein synthesis systems. Based on the obtained data, we have developed a system to optimize mRNA sequences. Using this system, we have achieved remarkable improvements in the synthesis yield of multiple proteins.
To date, RNA-based technologies are becoming increasingly important in the medical and biotechnology industries, and we are conducting research at the forefront of this field.
-
Figure 1: Process of gene expression
-
Figure 2: Selective transcription and mRNA diversity
The diversity of mRNA transcribed from a single gene is determined by the selection of transcription start sites, splicing patterns, and polyadenylation sites. Dring this process, mRNA sequences that encode different protein isoforms or vary in stability and translation efficiency are generated, allowing for intricate gene expression regulation. -
Figure 3: Development of an mRNA sequence optimization system
By utilizing sequencing technologies and machine learning, the key factors influencing mRNA stability and translation efficiency are identified to develop an mRNA sequence optimization system. This system aims to improve the effectiveness of mRNA therapeutics and enhance the efficiency of valuable protein production on a large scale.
Staff
- Assoc. Prof.: Shotaro Yamasaki
- SA Asst. Prof.: NEVIEN KAMEL MOHAMED ABDELKHALEK
Publications
1.Sequence features around cleavage sites are highly conserved among different species and a critical determinant for RNA cleavage position across eukaryotes. Ueno D., et al., J. Biosci. Bioeng. (2022) 143: 450-461
2.Methods for detecting RNA degradation intermediates in plants. Ueno D., et al., Plant Sci. (2022) 318: 111241
3.Feature selection for RNA cleavage efficiency at specific sites using the LASSO regression model in Arabidopsis thaliana. Ueno D., et al., BMC Bioinformatics (2022) 22: 380
4.Identification of 5'-untranslated regions that function as effective translational enhancers in monocotyledonous plant cells using a novel method of genome-wide analysis. Yamasaki, S., et al., Plant Biotechnol. (2018) 35: 365-373
5.Arabidopsis thaliana cold-regulated 47 gene 5-untranslated region enables stable high-level expression of transgenes. Yamasaki, S., et al., J. Biosci. Bioeng. (2018) 125: 124-130
6.Changes in Polysome Association of mRNA Throughout Growth and Development in Arabidopsis thaliana. Yamasaki, S., et al., Plant Cell Physiol. (2015) 56: 2169-2180
- Home
- Laboratories
- Yamasaki Lab