The main focus of this department is to elucidate the molecular mechanisms of viral diseases, including human immunodeficiency virus (HIV). The following projects are currently underway.
(1) Antiretroviral factors
HIV does not establish a productive infection in any monkey other than the chimpanzee. This is thought to be due to the fact that inhibitors in simian lymphocytes act at the early stage (reverse transcription) of viral infection. To date, TRIM5α and TRIMCyp have been identified as such restriction factors. We have shown that differences in the amino acid sequences in the C-terminal domain of TRIM5α of different monkey species affect the species-specific restriction of retrovirus infection. We also found that sequence variations in the N-terminal half of the viral capsid protein determine viral sensitivity to TRIM5α-mediated restriction, which indicates that there is an interaction between TRIM5α and the virus capsid. In addition, we found that HIV-2 replication levels in infected individuals associate with capsid variations. Furthermore, a single nucleotide polymorphism in the human TRIM5α gene appears to affect the rate of HIV-1 infection. These results strongly suggest that TRIM5α acts as an anti-HIV factor in HIV-infected individuals, which raises the possibility of a novel anti-HIV strategy based on potentiating TRIM5α.
(2) Host factors that participate in the adverse effects of anti-retroviral drugs
Human genetic variations drive differences in human phenotypes. In collaboration with Thai groups, we are analyzing the relationship between human genomic variation and adverse effects of antiretroviral therapy, with the aim of establishing “tailor-made therapies” that will improve the quality of life of HIV-infected patients.
(3) Analysis of HIV-1 genome RNA dimerization
The genome of a retrovirus such as HIV-1 is a single-stranded, positive-sense RNA. The viral genome always exists as a dimer in virions. Genome dimerization plays important roles in various stages of the viral life cycle, including genome packaging and reverse transcription, and in the genome recombination processes that are involved in viral diversification. We have constructed HIV-1 Gag cleavage site mutants and are assessing the effect of these mutations on the virion maturation steps of Gag processing, RNA dimerization, virion morphology, and infectivity by steady-state observation. We found that the maturation of the viral RNA/protein plays a critical role in the acquisition of viral infectivity (Fig. 2A). We previously identified the region that is necessary and sufficient for HIV-1 genome dimerization in the virion (DLS: Dimer Linkage Structure). By further fine mapping of the DLS, we discovered the possibility of a long-range interaction, which has never been reported previously. We performed computer-assisted structural modeling and obtained new 3D models of the HIV-1 DLS that revealed a unique pseudoknot-like conformation (Fig. 2B). Since this conformation appears to be thermodynamically stable, forms a foundational skeleton for the DLS, and sterically restricts the spontaneous diversification of DLS conformations, its unique shape may potentially serve as a novel target for anti-HIV-1 therapies.
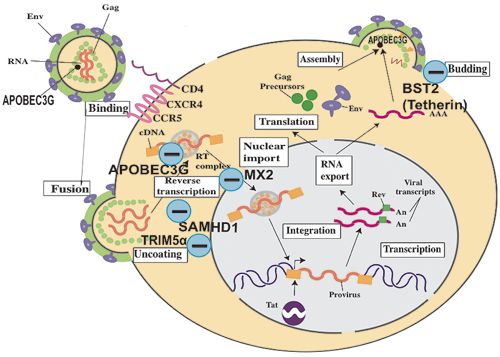
Fig. 1 Host factors that affect HIV replication. (-) indicates a negative regulator.
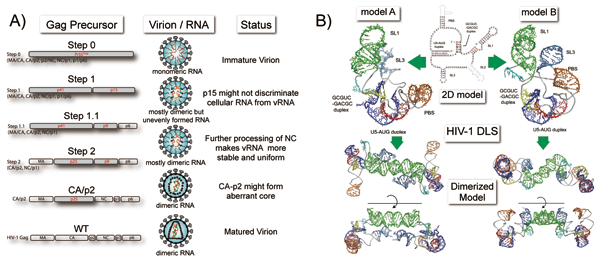
Fig. 2 HIV-1 genome RNA dimerization.