Positive-strand RNA viruses can dramatically rearrange the intracellular membranes of the host cell and produce unusual organelle-like structures called ‘replication complexes’ or ‘membranous webs’. These membrane structures appear in close proximity to the endoplasmic reticulum, and probably serve as a scaffold for the assembly of virus replication machinery by providing an organization and an environment that facilitate viral propagation. These structures are also known to serve as shells that protect the viruses against various cellular stress responses and allow persistent viral replication in the cytoplasm. Our group has been focusing on the molecular mechanisms involved in the formation of these replication complexes to determine the dynamic state of viral and/or host factors during the viral propagation cycle. Recently, we succeeded in purifying replication complexes from cells infected with hepatitis C virus or flavivirus. Quantitative mass spectrometry analyses led to the identification of several cellular factors that are specifically recruited to these viral replication complexes. We are currently focusing on the molecular functions of these newly identified cellular factors on the presumption that they are involved in the biosynthesis of the viral replication complexes. We are also studying the molecular mechanisms of viral particle formation and the mechanisms of autophagy induction seen in virus-infected cells. These studies may contribute to the development of novel antiviral therapies.
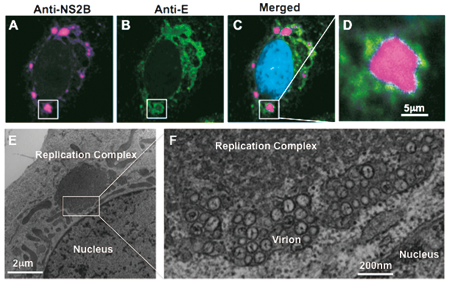
Figure: Fluorescent (A-D) and electron (E and F) microscope images of viral replication complexes in flavivirus-infected Vero cells. Twenty-four hours after infection, the cells were fixed and stained with antibodies against viral non-structural (anti-NS2B, magenta), structural (anti-E, green), and nuclear (DAPI, cyan) proteins. Unusual membrane structures, 5 to 10 nm in diameter and positive for viral antigens, are detected adjacent to the nucleus in the virus-infected cells. The endoplasmic reticulum is observed at the periphery of this structure, and many virus-like particles are detected inside of the lumen.