1. RAPID (Robotics Assisted Pathogen IDentification)
Under the aegis of the Program of Research Centers for Emerging and Re-emerging Infectious Diseases of MEXT, Japan, and in collaboration with the Omics Science Center, RIKEN, we are constructing a framework called “RAPID” that will facilitate the emergency diagnosis of infectious diseases. We are also cooperating with the research centers of eight countries in Asia and Africa in an effort to identify the causative agents in naturally occurring outbreaks.
2. Metagenomic Diagnosis of Infectious Diseases
Metagenomic analysis allows us to diagnose many of the major human infectious diseases (including respiratory tract infections, enteric infections, and blood-borne infections) by using a single common protocol. In addition, to pre-empt zoonotic disease outbreaks, we are seeking to identify new pathogenic microorganisms in animal-derived samples that may have zoonotic potential.
3. Metagenomic Analysis of the Intestinal Microbiome
The intestinal microbiome plays an important role in protecting the host from pathogen invasion. We are currently analyzing the intestinal microbiome of patients with diarrheal diseases to elucidate how the human host, the intestinal microbiome, and pathogenic microorganisms interact. This analysis will help us to understand the changes that occur in the intestinal microbiome during the course of infection.
4. Development of Novel Methods for Pathogen Detection
To develop more efficient and comprehensive methods of identifying pathogens, we are studying the efficacy of different methods for amplifying the genome of pathogenic microorganisms and subtracting the host genome.
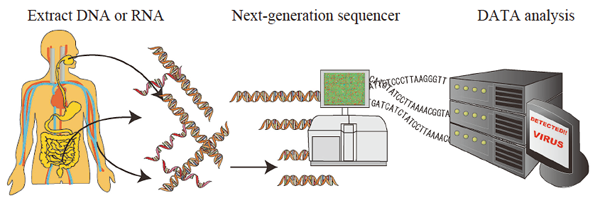
Fig. 1. Metagenomic diagnosis of infectious diseases using a next-generation sequencer.

Fig. 2. The next-generation sequencers installed in our department, Roche 454 GS Junior Bench Top System,
illumina MiSeq Personal Sequencer, Pacific Biosciences RS System.
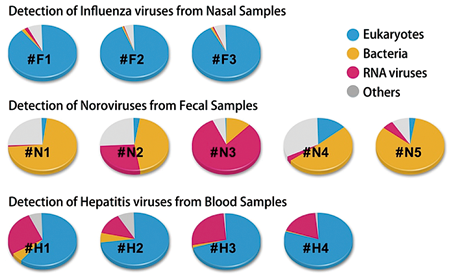
Fig. 3. Distribution of detected organisms in viral infection cases |
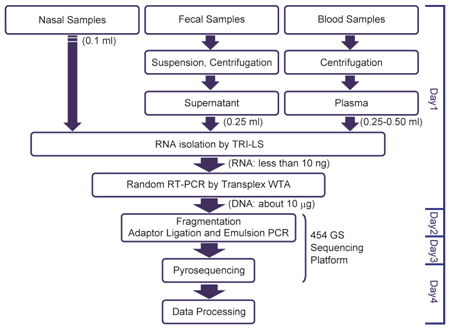
Fig. 4. Standard operating protocol for RAPID |