Research Group
| Associate Professor | Hodaka Fujii |
| Assistant Professor | Toshitsugu Fujita |
Research Projects
Locus-specific biochemical analysis of genome functions
A comprehensive understanding of the mechanisms behind genome functions such as transcription and epigenetic regulation requires the identification of the molecules that bind to the genomic regions of interest in vivo. However, there are few non-biased methods for identifying such molecules. To facilitate the biochemical and molecular biological analysis of specific genomic regions, we developed the locus-specific chromatin immunoprecipitation (ChIP) technologies to purify the genomic regions of interest. The ChIP technologies consist of insertional ChIP (iChIP) and engineered DNA-binding molecule-mediated ChIP (enChIP).
The scheme of iChIP is as follows (left panel): (i) The recognition sequence(s) of an exogenous DNA-binding protein such as LexA are inserted into the genomic region of interest in the cell that is to be analyzed. (ii) The DNA-binding domain (DNA DB) of the exogenous DNA-binding protein is fused with a tag(s) and a nuclear localization signal(s), and then expressed in the cell that is to be analyzed. (iii) The resulting cell is stimulated and crosslinked with formaldehyde or other crosslinkers, if necessary. (iv) The cell is lysed and the crosslinked DNA is fragmented by sonication. (v) The complexes, including the complexes bearing exogenous DNA DB, are immunoprecipitated with an antibody against the tag.
A comprehensive understanding of the mechanisms behind genome functions such as transcription and epigenetic regulation requires the identification of the molecules that bind to the genomic regions of interest in vivo. However, there are few non-biased methods for identifying such molecules. To facilitate the biochemical and molecular biological analysis of specific genomic regions, we developed the locus-specific chromatin immunoprecipitation (ChIP) technologies to purify the genomic regions of interest. The ChIP technologies consist of insertional ChIP (iChIP) and engineered DNA-binding molecule-mediated ChIP (enChIP).
The scheme of iChIP is as follows (left panel): (i) The recognition sequence(s) of an exogenous DNA-binding protein such as LexA are inserted into the genomic region of interest in the cell that is to be analyzed. (ii) The DNA-binding domain (DNA DB) of the exogenous DNA-binding protein is fused with a tag(s) and a nuclear localization signal(s), and then expressed in the cell that is to be analyzed. (iii) The resulting cell is stimulated and crosslinked with formaldehyde or other crosslinkers, if necessary. (iv) The cell is lysed and the crosslinked DNA is fragmented by sonication. (v) The complexes, including the complexes bearing exogenous DNA DB, are immunoprecipitated with an antibody against the tag.
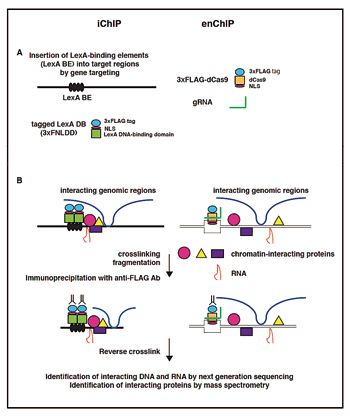
Figure. A scheme of locus-specific ChIP consisting of iChIP and enChIP
The scheme of enChIP is as follows (right panel): (i) A DNA-binding molecule/complex (DB) that recognizes the DNA sequence in a genomic region of interest is generated. This can be achieved by utilizing technologies such as engineered zinc-finger proteins, transcription activator-like (TAL) proteins, and the CRISPR system consisting of dCas9 together with gRNA. The engineered DB is fused with a tag(s) and an NLS(s), and then expressed in the cell that is to be analyzed. (ii) The resulting cell is stimulated and crosslinked with formaldehyde or other crosslinkers, if necessary. (iii) The cell is lysed, and the DNA is fragmented by sonication or digested with nucleases such as restriction enzymes. (iv) The complexes, including those containing the engineered DB, are subjected to affinity purification methods such as immunoprecipitation.
Thus, the complexes isolated by iChIP and enChIP retain molecules interacting with the genomic region of interest. Reverse crosslinking and subsequent purification of DNA, RNA, proteins, or other molecules allow these molecules to be identified and characterized.
We will apply locus-specific ChIP to elucidate the molecular mechanisms behind important epigenetic phenomena, including: (a) the lineage commitment of lymphocytes, (b) the regulation of odorant receptor expression, (c) the insulator function, (d) the detection and repair of DNA double-strand break, and (e) the epigenetic suppression of the expression of tumor suppressor genes.
Thus, the complexes isolated by iChIP and enChIP retain molecules interacting with the genomic region of interest. Reverse crosslinking and subsequent purification of DNA, RNA, proteins, or other molecules allow these molecules to be identified and characterized.
We will apply locus-specific ChIP to elucidate the molecular mechanisms behind important epigenetic phenomena, including: (a) the lineage commitment of lymphocytes, (b) the regulation of odorant receptor expression, (c) the insulator function, (d) the detection and repair of DNA double-strand break, and (e) the epigenetic suppression of the expression of tumor suppressor genes.
Major publications
- Fujita T, Asano Y, Ohtsuka J, Takada Y, Saito K, Ohki R, Fujii H. Identification of telomere-associated molecules by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP).Sci Rep. 2013 Nov 8;3:3171.
- Fujita T, Fujii H. Efficient isolation of specific genomic regions and identification of associated proteins by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP) using CRISPR. Biochem Biophys Res Commun. 2013 Sep 13;439(1):132-6.
- Fujita T, Fujii H. Direct identification of insulator components by insertional chromatin immunoprecipitation. PLoS One. 2011;6(10):e26109
- Hoshino A, Fujii H. Insertional chromatin immunoprecipitation: a method for isolating specific genomic regions. J Biosci Bioeng. 2009 Nov;108(5):446-9.
- Saint Fleur S, Hoshino A, Kondo K, Egawa T, Fujii H. Regulation of Fas-mediated immune homeostasis by an activation-induced protein, Cyclon. Blood. 2009 Aug 13;114(7):1355-65.